12/10/2008-
Small bubbles had accumulated along the edges of all 4 strips, despite allowing strips and reagents to warm to RT before loading the cassette. Cassette was tapped vigorously to try to get bubbles out, but wasn't successful.
IEF was performed on the 4 strips from yesterday with the following parameters:
250V - 20 mins.
450V - 15 mins.
750V - 15 mins.
2000V - 60 mins.
The run looked decent. Got some "rippling" at the "-" end of the strip towards the end of the run, but that has been the norm at this pont.
Strips were washed with 10mL reducing buffer for 15mins and then 10mL of alkylating buffer for 15mins. Strips were transferred to ZOOM 4-20% tris-glycine gels and overlayed with 0.5% agarose solution made with tris-glycine buffer. Gels were run @ 125V for ~2.5 hrs until dye front had reached the end of the gels.
NOTE: Samples #1 and #2 were accidentally not connected properly to the power supply (lid was on wrong) and did not receive any current for ~1hr. Lid was adjusted and the gel was run out. Looked normal.
Gels were stained by Tatyana using the fast protocol and then stored O/N @ 4C.
Gel #1 - Sterile sea H2O+V.tubiashii -
Gel #2 - V.tubiashii + Live C.gigas -
Gel #3 - V.tubiashii + Autoclaved C.gigas -
Gel #4 - Sterile sea H2O + Live C.gigas -
Notes: In gels #3 and #4 it looks like the ladder leaked over into the sample well as can be seen by the horizontal streaking that comes from each band of the ladder.
12/09/2008
Quantification of the protein in the sample from 12/08/08
Samples were desalted today by Sam
Procedure for quantification:
1. Mix 5 microL of sample with 250 microL of Coomassie reagent
2. Incubated at RT for 10 min
3. Measure Abs @ 595nm
Results:
| VT+live oysters |
VT+ autoclaved oysters |
VT+H2O |
Live oysters in sea H2O |
|
| 0.021 |
0.035 |
0.017 |
0.000 |
|
| 0.022 |
0.041 |
0.010 |
0.000 |
|
| 0.023 |
0.033 |
0.022 |
0.000 |
|
| 0.020 |
0.037 |
0.020 |
0.000 |
|
| 0.023 |
0.038 |
0.013 |
0.000 |
|
| Average |
0.022 |
0.0368 |
0.0164 |
0.000 |
C= ((Aave -0.04) + 0.0403)/0.00007
VT+ Alive:
C =((0.022-0.04)+0.0403)/0.00007 = 318.57 microg/mL
VT + autoclaved
C= ((0.0368-0.04) + 0.0403)/0.00007 = 530 microg/mL
VT +H2O
C = (( 0.0164- 0.04)- 0.0403)/0.00007 = 238.6 microg/mL
Gigas H2O
C = 0 microg/mL
AMOUNT OF THE PROTEIN USED IN THE LAST 2-D GELL RUN = 0.623 MICROG
VT + alive
Volume = 0.623 * 1000 / 318.57 = 1.955 microL
VT + autoclaved
Volume = 0.623 * 1000/530 = 1.17 microL
VT +H2O
Volume = 0.623 *1000/238.6 = 2.611 microL
Oysters H2O
Voulme = 5 microL
Rehydration Buffer Master Mix
1. 2-D solubilizer #2
141 *5
705 microL2. 2M DTT = 0.775 *5 = 3.875 microL
3. Carreir Ampholytes = 0.775 * 5 = 3.875 microL
4. Bromophenol Blue = 0.1 *5 = 0.5 microL
2M DTT was prepaired by adding 0.0308 g of the DTT (MW = 154.2 g) to 100 microL of H2O
142.62 microL of the MM was added to each sample. H2O was added to each sample to get volume to 155 microL.
155 microL of the each sample was loaded into sampe loading wells, IPG strips were placed into the well with the gel side up
Sample to let at RT overnight for rehydration
12/08/08
Isolation of Proteins for 2-D Gel
Sample are from Exposure Experiment, t=2
1. sterile H2O + VT
2. Alive oysters + sea H2O + VT
3. Autoclaved oysters + sterile H2O + VT
4. Alive Oysters + sea H2O
Procedure:
1. Each pellet was thawed and resuspended in 500 microL of Lysis buffer (pBAD)
2. Sonicator was used to isolate proteins
3. Samples were centrifuged at max speed for 1 min at +4 C to pellet insoluble proteins.
4. Supernates were transfered into clean tubes and placed in the -80C freezer.
12/02/2008

1/3 Anoxic, 6hours
1.
My estimation (5.6,57)
Results: GPPA_VIBVY (Q7MGP6)
Guanosine-5'-triphosphate,3'-diphosphate pyrophosphatase.
Chain: 1-497, pI: 5.55, Mw: 54755
GLGA_VIBVY (Q7MJ50)
Glycogen synthase.
Chain: 1-485, pI: 5.55, Mw: 54229
GPPA_VIBVU (Q8DDN5)
Guanosine-5'-triphosphate,3'-diphosphate pyrophosphatase.
Chain: 1-497, pI: 5.55, Mw: 54755
RF3_VIBCH (Q9KU64)
Peptide chain release factor 3.
Chain: 1-531, pI: 5.59, Mw: 59626
2.
My estimation (5.52, 89)
Results: GYRB_VIBPA (O51859)
DNA gyrase subunit B.
Chain: 1-805, pI: 5.53, Mw: 89443
3.
My estimation (5.52, 81)
Results: FADB_VIBPA (Q87TN9)
Fatty acid oxidation complex subunit alpha.
Chain: 1-723, pI: 5.53, Mw: 78556
4.
My estimation (5.6,91)
Results: HEX_VIBVU (Q04786)
Beta-hexosaminidase.
Chain: 1-847, pI: 5.63, Mw: 94277
MUTS_VIBVY (Q7MHR2)
DNA mismatch repair protein mutS.
Chain: 1-853, pI: 5.70, Mw: 94485
MUTS_VIBVU (Q8DC53)
DNA mismatch repair protein mutS.
Chain: 1-853, pI: 5.70, Mw: 94483
PBPA_VIBCH (Q9KNU5)
Penicillin-binding protein 1A.
Chain: 1-825, pI: 5.63, Mw: 91987
GYRB_VIBCH (Q9KVX3)
DNA gyrase subunit B.
Chain: 1-805, pI: 5.69, Mw: 89520
5.
My estimation (5.6; 89)
Results: PBPA_VIBCH (Q9KNU5)
Penicillin-binding protein 1A.
Chain: 1-825, pI: 5.63, Mw: 91987
GYRB_VIBCH (Q9KVX3)
DNA gyrase subunit B.
Chain: 1-805, pI: 5.69, Mw: 89520
6.
My estimation: (5.6;85)
METE_VIBVY (Q7MJM6)
5-methyltetrahydropteroyltriglutamate-- homocysteine meth...
Chain: 1-778, pI: 5.55, Mw: 86960
7.
My estimation (6; 100) and (6;92?)- two bands are close to each other
UVRA_VIBVY (Q7MHB5)
UvrABC system protein A.
Chain: 1-940, pI: 6.12, Mw: 103995
UVRA_VIBPA (Q87LA0)
UvrABC system protein A.
Chain: 1-940, pI: 6.11, Mw: 103872
UVRA_VIBVU (Q8DCJ3)
UvrABC system protein A.
Chain: 1-940, pI: 6.08, Mw: 103961
MALT_VIBCH (Q9KNF3)
HTH-type transcriptional regulator malT.
Chain: 1-902, pI: 6.00, Mw: 103589
UVRA_VIBCH (Q9KUW5)
UvrABC system protein A.
Chain: 1-940, pI: 6.08, Mw: 104328
8.
My estimation : three proteins with same Isoelectric point of 5.9 aand MW: 120,110,105
Results:
TAGA_VIBC3 (A5F398)
ToxR-activated gene A lipoprotein.
Chain: 22-1002, pI: 5.96, Mw: 112264
TAGA_VIBCH (P0C6Q7)
ToxR-activated gene A lipoprotein.
Chain: 22-1002, pI: 5.96, Mw: 112248
MALT_VIBCH (Q9KNF3)
HTH-type transcriptional regulator malT.
Chain: 1-902, pI: 6.00, Mw: 103589
9.
My estimation: two proteins 6.05; MW 122 and 110
UVRA_VIBVY (Q7MHB5)
UvrABC system protein A.
Chain: 1-940, pI: 6.12, Mw: 103995
UVRA_VIBPA (Q87LA0)
UvrABC system protein A.
Chain: 1-940, pI: 6.11, Mw: 103872
UVRA_VIBVU (Q8DCJ3)
UvrABC system protein A.
Chain: 1-940, pI: 6.08, Mw: 103961
UVRA_VIBCH (Q9KUW5)
UvrABC system protein A.
Chain: 1-940, pI: 6.08, Mw: 104328
10.
My estimation: 6.1, 81
MNMG_VIBPA (Q87K98)
tRNA uridine 5-carboxymethylaminomethyl modification enzy...
Chain: 1-631, pI: 6.07, Mw: 70060
MNMC_VIBCH (Q9KQ91)
tRNA 5-methylaminomethyl-2-thiouridine biosynthesis bifun...
Chain: 1-674, pI: 6.08, Mw: 74791
11.
My estimation (5.9, 70)
SYT_VIBC3 (A5EZ18)
Threonyl-tRNA synthetase.
Chain: 1-642, pI: 5.95, Mw: 73453
DXS_VIBC3 (A5F331)
1-deoxy-D-xylulose-5-phosphate synthase.
Chain: 1-626, pI: 5.86, Mw: 68347
MNMG_VIBVY (Q7MGG9)
tRNA uridine 5-carboxymethylaminomethyl modification enzy...
Chain: 1-632, pI: 5.92, Mw: 70205
MACB_VIBPA (Q87JM4)
Macrolide export ATP-binding/permease protein macB.
Chain: 1-654, pI: 5.86, Mw: 71213
DXS_VIBPA (Q87RU0)
1-deoxy-D-xylulose-5-phosphate synthase.
Chain: 1-621, pI: 5.82, Mw: 67849
MNMG_VIBVU (Q8DDH9)
tRNA uridine 5-carboxymethylaminomethyl modification enzy...
Chain: 1-632, pI: 5.92, Mw: 70189
SYT_VIBCH (Q9KMN7)
Threonyl-tRNA synthetase.
Chain: 1-642, pI: 5.92, Mw: 73444
DXS_VIBCH (Q9KTL3)
1-deoxy-D-xylulose-5-phosphate synthase.
Chain: 1-626, pI: 5.86, Mw: 68347
ACSA_VIBCH (Q9KV59)
Acetyl-coenzyme A synthetase.
Chain: 1-649, pI: 5.94, Mw: 71895
12. My estimation (5.9; 64)
HUTU_VIBC3 (A5F1X6)
Urocanate hydratase.
Chain: 1-565, pI: 5.91, Mw: 61825
BETA_VIBHB (A7N2P9)
Choline dehydrogenase.
Chain: 1-569, pI: 5.92, Mw: 62530
BETA_VIBVY (Q7MF12)
Choline dehydrogenase.
Chain: 1-560, pI: 5.92, Mw: 62360
MAO11_VIBVY (Q7MLG3)
NAD-dependent malic enzyme 1.
Chain: 1-562, pI: 6.00, Mw: 62089
MEND_VIBVY (Q7MMF6)
2-succinyl-5-enolpyruvyl-6-hydroxy-3- cyclohexene-1-carbo...
Chain: 1-564, pI: 5.96, Mw: 61826
FTHS_VIBPA (Q87HX2)
Formate--tetrahydrofolate ligase.
Chain: 1-582, pI: 5.91, Mw: 62338
BETA_VIBVU (Q8D3K2)
Choline dehydrogenase.
Chain: 1-560, pI: 5.96, Mw: 62278
MEND_VIBVU (Q8D820)
2-succinyl-5-enolpyruvyl-6-hydroxy-3- cyclohexene-1-carbo...
Chain: 1-564, pI: 5.91, Mw: 61870
MAO1_VIBVU (Q8D911)
NAD-dependent malic enzyme.
Chain: 1-562, pI: 6.00, Mw: 62089
HUTU_VIBCH (Q9KSQ3)
Urocanate hydratase.
Chain: 1-565, pI: 5.91, Mw: 61825
13.
My estimation: (6.1; 60)
VIBE_VIBCH (O07899)
Vibriobactin-specific 2,3- dihydroxybenzoate-AMP ligase.
Chain: 1-543, pI: 6.11, Mw: 60066
LUXO_VIBC3 (O87455)
Regulatory protein luxO.
Chain: 1-530, pI: 6.15, Mw: 59027
MAO11_VIBVY (Q7MLG3)
NAD-dependent malic enzyme 1.
Chain: 1-562, pI: 6.00, Mw: 62089
MAO1_VIBVU (Q8D911)
NAD-dependent malic enzyme.
Chain: 1-562, pI: 6.00, Mw: 62089
PYRG_VIBCH (Q9KPC4)
CTP synthase.
Chain: 2-545, pI: 6.09, Mw: 59755
14.
(6.05;53)
THII_VIBVY (Q7MN44)
Thiamine biosynthesis protein thiI.
Chain: 1-482, pI: 6.04, Mw: 54720
ENGA_VIBVY (Q7MNE7)
GTP-binding protein engA.
Chain: 1-496, pI: 6.01, Mw: 55485
STHA_VIBPA (Q87KN5)
Soluble pyridine nucleotide transhydrogenase.
Chain: 1-466, pI: 6.01, Mw: 51349
THII_VIBPA (Q87RT6)
Thiamine biosynthesis protein thiI.
Chain: 1-482, pI: 6.04, Mw: 54632
15.
(5.85; 34)
END4_VIBHB (A7MS84)
Probable endonuclease 4.
Chain: 1-295, pI: 5.87, Mw: 33084
END4_VIBVY (Q7MNR1)
Probable endonuclease 4.
Chain: 1-293, pI: 5.86, Mw: 32726
RP32_VIBVU (Q8DD54)
RNA polymerase sigma-32 factor.
Chain: 1-285, pI: 5.88, Mw: 32326
END4_VIBVU (Q8DEP6)
Probable endonuclease 4.
Chain: 1-293, pI: 5.86, Mw: 32679
PPNK_VIBVU (Q8DF58)
Probable inorganic polyphosphate/ATP-NAD kinase.
Chain: 1-294, pI: 5.88, Mw: 32555
MRAW_VIBPA (Q9AJH1)
S-adenosyl-L-methionine-dependent methyltransferase mraW.
Chain: 1-316, pI: 5.88, Mw: 35214
Y3667_VIBHB (Q9KIQ6)
UPF0042 protein VIBHAR_03667.
Chain: 1-287, pI: 5.88, Mw: 32416
16.
(6.1,31)
FADR_VIBC3 (A5F6Z2)
Fatty acid metabolism regulator protein.
Chain: 1-279, pI: 6.11, Mw: 31998
ATP6_VIBAL (P12984)
ATP synthase a chain.
Chain: 1-270, pI: 6.17, Mw: 30134
LIP_VIBCH (P15493)
Lipase.
Chain: 23-312, pI: 6.11, Mw: 30648
FADR_VIBCH (Q9KQU8)
Fatty acid metabolism regulator protein.
Chain: 1-279, pI: 6.11, Mw: 31998
17.
(5.85;31)
Y252_VIBC3 (A5F0Z4)
Putative phosphotransferase VC0395_0252.
Chain: 1-277, pI: 5.86, Mw: 31802
KDSA_VIBC3 (A5F692)
2-dehydro-3-deoxyphosphooctonate aldolase.
Chain: 1-283, pI: 5.86, Mw: 30774
DAPF_VIBHB (A7N0W0)
Diaminopimelate epimerase.
Chain: 1-276, pI: 5.86, Mw: 30429
NAGB_VIBVY (Q7MGE1)
Glucosamine-6-phosphate deaminase.
Chain: 1-266, pI: 5.86, Mw: 29662
NAGB_VIBPA (Q87K60)
Glucosamine-6-phosphate deaminase.
Chain: 1-266, pI: 5.86, Mw: 29658
NAGB_VIBVU (Q8D4T9)
Glucosamine-6-phosphate deaminase.
Chain: 1-266, pI: 5.86, Mw: 29662
NANK_VIBVU (Q8D612)
N-acetylmannosamine kinase.
Chain: 1-293, pI: 5.82, Mw: 30969
Y1880_VIBVU (Q8DBE1)
UPF0294 protein VV1_1880.
Chain: 1-284, pI: 5.82, Mw: 31855
Y3786_VIBCH (Q9KKW4)
Putative phosphotransferase VC_A0986.
Chain: 1-277, pI: 5.86, Mw: 31802
PANE_VIBCH (Q9KPQ9)
2-dehydropantoate 2-reductase.
Chain: 1-296, pI: 5.82, Mw: 32201
KDSA_VIBCH (Q9KQ29)
2-dehydro-3-deoxyphosphooctonate aldolase.
Chain: 1-283, pI: 5.86, Mw: 30774
18.
(6.2;107) and (6.2;105)
UVRA_VIBVY (Q7MHB5)
UvrABC system protein A.
Chain: 1-940, pI: 6.12, Mw: 103995
UVRA_VIBPA (Q87LA0)
UvrABC system protein A.
Chain: 1-940, pI: 6.11, Mw: 103872
19.
(6.2;98) and (6.2;92)
Same proteins as previous entry (#18)
20.
(5.7;38)
GCP_VIBC3 (A5F9E8)
Probable O-sialoglycoprotein endopeptidase.
Chain: 1-339, pI: 5.71, Mw: 36507
HUTG_VIBVY (P60111)
Formimidoylglutamase.
Chain: 1-336, pI: 5.70, Mw: 37499
GPDA_VIBVY (Q7MGY7)
Glycerol-3-phosphate dehydrogenase [NAD(P)+].
Chain: 1-345, pI: 5.75, Mw: 36961
TRUD_VIBVY (Q7MHQ6)
tRNA pseudouridine synthase D.
Chain: 1-347, pI: 5.71, Mw: 38524
META_VIBVY (Q7MLD5)
Homoserine O-succinyltransferase.
Chain: 1-313, pI: 5.75, Mw: 36377
ASTE_VIBVY (Q7MLE8)
Succinylglutamate desuccinylase.
Chain: 1-342, pI: 5.71, Mw: 38847
RSGA2_VIBPA (Q87FP9)
Putative ribosome biogenesis GTPase rsgA 2.
Chain: 1-358, pI: 5.71, Mw: 39803
HUTG_VIBVU (Q8DA19)
Formimidoylglutamase.
Chain: 1-336, pI: 5.70, Mw: 37515
TRUD_VIBVU (Q8DC58)
tRNA pseudouridine synthase D.
Chain: 1-347, pI: 5.71, Mw: 38554
GPDA_VIBVU (Q8DCW4)
Glycerol-3-phosphate dehydrogenase [NAD(P)+].
Chain: 1-345, pI: 5.75, Mw: 36961
21.
(5.5;60)
SCRB_VIBC3 (A5EZZ8)
Probable sucrose-6-phosphate hydrolase.
Chain: 1-546, pI: 5.50, Mw: 62313
RECN_VIBC3 (A5F379)
DNA repair protein recN.
Chain: 1-554, pI: 5.50, Mw: 61252
HUTU_VIBHB (A7MVK1)
Urocanate hydratase.
Chain: 1-565, pI: 5.50, Mw: 62123
RECN_VIBCH (P0C6Q4)
DNA repair protein recN.
Chain: 1-554, pI: 5.50, Mw: 61252
DSBD_VIBCH (Q9KNN1)
Thiol:disulfide interchange protein dsbD.
Chain: 22-600, pI: 5.49, Mw: 62819
22. (4.8, 18)
Y3078_VIBHB (A7MUF3)
UPF0260 protein VIBHAR_03078.
Chain: 1-150, pI: 4.80, Mw: 17432
HLY2_VIBPA (P19250)
Thermostable direct hemolysin 2.
Chain: 25-189, pI: 4.79, Mw: 18670
DEF2_VIBPA (Q87I22)
Peptide deformylase 2.
Chain: 1-168, pI: 4.79, Mw: 18491
23.
(5.5; 17)
FUR_VIBVU (P33117)
Ferric uptake regulation protein.
Chain: 1-149, pI: 5.51, Mw: 16743
\

1/3 Aerobic, 6 hours
1.
(5.7, 114)
SBCC_VIBCH (Q9KM67)
Nuclease sbcCD subunit C.
Chain: 1-1013, pI: 5.71, Mw: 114593
2.
(5.75,130)
Autoinducer 2 sensor kinase/phosphatase luxQ.
Chain: 1-857, pI: 5.74, Mw: 96930
3.
(4.5, 81)
COLA_VIBPA (Q56696)
Microbial collagenase.
Chain: 28-816, pI: 4.57, Mw: 88379
4.
(5.6,114)
GLNE_VIBVY (Q7MNY4)
Glutamate-ammonia-ligase adenylyltransferase.
Chain: 1-950, pI: 5.58, Mw: 108919
GLNE_VIBVU (Q8DEH6)
Glutamate-ammonia-ligase adenylyltransferase.
Chain: 1-950, pI: 5.58, Mw: 109007
5.
(4.6,64)
HSCA_VIBHB (A7MU49)
Chaperone protein hscA homolog.
Chain: 1-617, pI: 4.62, Mw: 66100
BTUB_VIBVU (Q8DD41)
Vitamin B12 transporter btuB.
Chain: 23-606, pI: 4.61, Mw: 64354
6.
(5.3,132)
DPO3A_VIBCH (P52022)
DNA polymerase III subunit alpha.
Chain: 1-1159, pI: 5.20, Mw: 130058
7.
(5.1,81)
SYM_VIBHB (A7MZT3)
Methionyl-tRNA synthetase.
Chain: 1-686, pI: 5.08, Mw: 77625
SYM_VIBVY (Q7MM14)
Methionyl-tRNA synthetase.
Chain: 1-690, pI: 5.12, Mw: 78120
UVRB_VIBPA (Q87MX6)
UvrABC system protein B.
Chain: 1-676, pI: 5.12, Mw: 77070
SYM_VIBPA (Q87N07)
Methionyl-tRNA synthetase.
Chain: 1-688, pI: 5.16, Mw: 77995
SYM_VIBVU (Q8D8F2)
Methionyl-tRNA synthetase.
Chain: 1-690, pI: 5.12, Mw: 78120
8.
(5,55)
SYE_VIBHB (A7MY61)
Glutamyl-tRNA synthetase.
Chain: 1-475, pI: 5.00, Mw: 53291
GUAA_VIBVY (Q7MNE1)
GMP synthase [glutamine-hydrolyzing].
Chain: 1-517, pI: 5.00, Mw: 57662
MLTF_VIBPA (Q87RW1)
Membrane-bound lytic murein transglycosylase F.
Chain: 25-525, pI: 5.00, Mw: 57090
GUAA_VIBVU (Q8DF07)
GMP synthase [glutamine-hydrolyzing].
Chain: 1-517, pI: 5.00, Mw: 57676
9.
(5.7;55)
ALDH_VIBC3 (A5F3A7)
Aldehyde dehydrogenase.
Chain: 1-506, pI: 5.66, Mw: 55885
ATPA_VIBC3 (A5F457)
ATP synthase subunit alpha.
Chain: 1-513, pI: 5.66, Mw: 55650
MURC_VIBC3 (A5F5M8)
UDP-N-acetylmuramate--L-alanine ligase.
Chain: 1-486, pI: 5.75, Mw: 53042
GPPA_VIBHB (A7MXT2)
Guanosine-5'-triphosphate,3'-diphosphate pyrophosphatase.
Chain: 1-497, pI: 5.69, Mw: 54724
ALDH_VIBCH (P0C6D7)
Aldehyde dehydrogenase.
Chain: 1-506, pI: 5.66, Mw: 55885
ATPA_VIBCH (Q9KNH3)
ATP synthase subunit alpha.
Chain: 1-513, pI: 5.66, Mw: 55650
MURC_VIBCH (Q9KPG8)
UDP-N-acetylmuramate--L-alanine ligase.
Chain: 1-486, pI: 5.75, Mw: 53042
10.
(5.7;51)
DLDH_VIBPA (O50286)
Dihydrolipoyl dehydrogenase.
Chain: 1-475, pI: 5.71, Mw: 50988
LUXO_VIBVY (Q7MM78)
Regulatory protein luxO.
Chain: 1-453, pI: 5.71, Mw: 50515
LEUC_VIBPA (Q87SS9)
3-isopropylmalate dehydratase large subunit.
Chain: 1-471, pI: 5.69, Mw: 50618
LUXO_VIBVU (Q8CWJ5)
Regulatory protein luxO.
Chain: 1-453, pI: 5.71, Mw: 50515
11.
(5.75,50)
LUXO_VIBPA (Q87MX7)
Regulatory protein luxO.
Chain: 1-453, pI: 5.71, Mw: 50482
NRFA_VIBPA (Q87ND9)
Cytochrome c-552.
Chain: 30-475, pI: 5.71, Mw: 50253
PNCB_VIBVU (Q8DA38)
Nicotinate phosphoribosyltransferase.
Chain: 1-437, pI: 5.69, Mw: 50002
12.
LPXB_VIBHB (A7MY02)
Lipid-A-disaccharide synthase.
Chain: 1-379, pI: 5.69, Mw: 42782
FABF_VIBCH (Q9KQH9)
3-oxoacyl-[acyl-carrier-protein] synthase 2.
Chain: 2-414, pI: 5.66, Mw: 43077
13.
(5.8;130)
SBCC_VIBCH (Q9KM67)
Nuclease sbcCD subunit C.
Chain: 1-1013, pI: 5.71, Mw: 114593
14.
(5.8;114)
MALT_VIBHB (A7N5N6)
HTH-type transcriptional regulator malT.
Chain: 1-902, pI: 5.78, Mw: 103870
15.
(5.85;131)
RPOC_VIBPA (Q87KQ5)
DNA-directed RNA polymerase subunit beta'.
Chain: 1-1400, pI: 5.82, Mw: 154920
GLNE_VIBCH (Q9KPD4)
Glutamate-ammonia-ligase adenylyltransferase.
Chain: 1-948, pI: 5.83, Mw: 109334
16.
(5.9;125)
GCSP_VIBC3 (A5EYY8)
Glycine dehydrogenase [decarboxylating].
Chain: 1-954, pI: 5.87, Mw: 103955
MALT_VIBVY (Q7MG94)
HTH-type transcriptional regulator malT.
Chain: 1-902, pI: 5.88, Mw: 103847
MALT_VIBPA (Q87FQ5)
HTH-type transcriptional regulator malT.
Chain: 1-902, pI: 5.88, Mw: 104078
MALT_VIBVU (Q8D4P3)
HTH-type transcriptional regulator malT.
Chain: 1-902, pI: 5.88, Mw: 103847
17.
(6;120)
TAGA_VIBC3 (A5F398)
ToxR-activated gene A lipoprotein.
Chain: 22-1002, pI: 5.96, Mw: 112264
TAGA_VIBCH (P0C6Q7)
ToxR-activated gene A lipoprotein.
Chain: 22-1002, pI: 5.96, Mw: 112248
18.
(6;106)
UVRA_VIBVU (Q8DCJ3)
UvrABC system protein A.
Chain: 1-940, pI: 6.08, Mw: 103961
MALT_VIBCH (Q9KNF3)
HTH-type transcriptional regulator malT.
Chain: 1-902, pI: 6.00, Mw: 103589
UVRA_VIBCH (Q9KUW5)
UvrABC system protein A.
Chain: 1-940, pI: 6.08, Mw: 104328
19.
(6.05,98)
CAPP_VIBCH (Q9KNT4)
Phosphoenolpyruvate carboxylase.
Chain: 1-876, pI: 5.92, Mw: 98317
20.
(6.15;58)
LUXO_VIBC3 (O87455)
Regulatory protein luxO.
Chain: 1-530, pI: 6.15, Mw: 59027
21.
(5.9;43)
LPXB_VIBVY (Q7MIH2)
Lipid-A-disaccharide synthase.
Chain: 1-380, pI: 5.91, Mw: 42610
LPXB_VIBVU (Q8DBE8)
Lipid-A-disaccharide synthase.
Chain: 1-380, pI: 5.91, Mw: 42610
Y1345_VIBCH (Q9KSB4)
Putative dioxygenase VC_1345.
Chain: 1-378, pI: 5.91, Mw: 43247
22.
(5.9;40)
E4PD_VIBPA (Q87LL0)
D-erythrose-4-phosphate dehydrogenase.
Chain: 1-345, pI: 5.91, Mw: 38250
23.
(5.6;36)
NAGZ_VIBC3 (A5F8Y1)
Beta-hexosaminidase.
Chain: 1-330, pI: 5.59, Mw: 36382
ARGC_VIBPA (Q87L55)
N-acetyl-gamma-glutamyl-phosphate reductase.
Chain: 1-334, pI: 5.61, Mw: 36177
Y1259_VIBCH (Q9KSJ7)
UPF0176 protein VC_1259.
Chain: 1-327, pI: 5.62, Mw: 37167
24.
(5.65;30)
PSTB2_VIBVY (Q7MFE8)
Phosphate import ATP-binding protein pstB 2.
Chain: 1-279, pI: 5.66, Mw: 31496
DAPF_VIBPA (Q87KJ4)
Diaminopimelate epimerase.
Chain: 1-276, pI: 5.66, Mw: 30315
MURI_VIBPA (Q87KP1)
Glutamate racemase.
Chain: 1-270, pI: 5.66, Mw: 29481
Y3769_VIBCH (Q9KKY1)
Pirin-like protein VC_A0969.
Chain: 1-282, pI: 5.66, Mw: 31325
25.
(5.7;29)
BIOH_VIBHB (A7MST3)
Carboxylesterase bioH.
Chain: 1-254, pI: 5.71, Mw: 28017
BTUD_VIBHB (A7MVV6)
Vitamin B12 import ATP-binding protein btuD.
Chain: 1-255, pI: 5.71, Mw: 27883
AROE_VIBHB (A7N127)
Shikimate dehydrogenase.
Chain: 1-277, pI: 5.71, Mw: 29954
XNI_VIBVY (Q7MN34)
Uncharacterized exonuclease xni.
Chain: 1-259, pI: 5.71, Mw: 29347
NQRC_VIBPA (Q87MA8)
Na(+)-translocating NADH-quinone reductase subunit C.
Chain: 1-261, pI: 5.71, Mw: 27690
26.
(5.5;33)
TAL_VIBCH (Q9KLW8)
Transaldolase.
Chain: 1-316, pI: 5.49, Mw: 34636
DAPA_VIBCH (Q9KQ47)
Dihydrodipicolinate synthase.
Chain: 1-292, pI: 5.50, Mw: 31374
27.
(5.5;27)
LFTR_VIBHB (A7N1L9)
Leucyl/phenylalanyl-tRNA--protein transferase.
Chain: 1-236, pI: 5.49, Mw: 26621
Y4273_VIBVU (Q8D788)
UPF0271 protein VV2_0273.
Chain: 1-247, pI: 5.50, Mw: 27363
Y2093_VIBCH (Q9KQA7)
UPF0135 protein VC_2093.
Chain: 1-252, pI: 5.50, Mw: 27868
28.
(5.3;60)
TRPE_VIBPA (P22099)
Anthranilate synthase component 1.
Chain: 1-541, pI: 5.30, Mw: 59771
G6PI_VIBPA (Q87L81)
Glucose-6-phosphate isomerase.
Chain: 1-550, pI: 5.28, Mw: 60913
NHAB_VIBPA (Q87N04)
Na(+)/H(+) antiporter nhaB.
Chain: 1-528, pI: 5.30, Mw: 57165
G6PI_VIBVU (Q8DCK7)
Glucose-6-phosphate isomerase.
Chain: 1-550, pI: 5.28, Mw: 60758
29.
(5.6;57)
NHAB_VIBC3 (A5F6Z3)
Na(+)/H(+) antiporter nhaB.
Chain: 1-530, pI: 5.61, Mw: 57662
NHAB_VIBCH (Q9KQU7)
Na(+)/H(+) antiporter nhaB.
Chain: 1-530, pI: 5.61, Mw: 57662
30.
(5.6;53)
PTYBC_VIBPA (Q87FD5)
PTS system N-acetylmuramic acid-specific EIIBC component.
Chain: 1-484, pI: 5.61, Mw: 51201
ARAA_VIBPA (Q87FK3)
L-arabinose isomerase.
Chain: 1-497, pI: 5.61, Mw: 54862
HUTH_VIBCH (Q9KSQ4)
Histidine ammonia-lyase.
Chain: 1-511, pI: 5.62, Mw: 54817
LUXO_VIBCH (Q9KT84)
Regulatory protein luxO.
Chain: 1-455, pI: 5.61, Mw: 50839
31.
(6;60)
G6PI_VIBC3 (A5F3J3)
Glucose-6-phosphate isomerase.
Chain: 1-550, pI: 5.59, Mw: 60692
PPCK_VIBC3 (A5F4Q4)
Phosphoenolpyruvate carboxykinase [ATP].
Chain: 1-542, pI: 5.61, Mw: 59844
PPCK_VIBCH (Q9KNK0)
Phosphoenolpyruvate carboxykinase [ATP].
Chain: 1-542, pI: 5.61, Mw: 59844
RF3_VIBCH (Q9KU64)
Peptide chain release factor 3.
Chain: 1-531, pI: 5.59, Mw: 59626
32.
(6;58)
NHAB_VIBC3 (A5F6Z3)
Na(+)/H(+) antiporter nhaB.
Chain: 1-530, pI: 5.61, Mw: 57662
NHAB_VIBCH (Q9KQU7)
Na(+)/H(+) antiporter nhaB.
Chain: 1-530, pI: 5.61, Mw: 57662
RF3_VIBCH (Q9KU64)
Peptide chain release factor 3.
Chain: 1-531, pI: 5.59, Mw: 59626
33.
(6;52)
PTYBC_VIBPA (Q87FD5)
PTS system N-acetylmuramic acid-specific EIIBC component.
Chain: 1-484, pI: 5.61, Mw: 51201
LUXO_VIBCH (Q9KT84)
Regulatory protein luxO.
Chain: 1-455, pI: 5.61, Mw: 50839
34.
(6.4;36)
ERA_VIBPA (Q87LP0)
GTP-binding protein era homolog.
Chain: 1-320, pI: 6.42, Mw: 36566
35.
(6.3;48)
Y2091_VIBVU (P59353)
UPF0229 protein VV1_2091.
Chain: 1-423, pI: 6.32, Mw: 48756
Y2350_VIBVY (Q7MJ13)
UPF0229 protein VV2350.
Chain: 1-423, pI: 6.32, Mw: 48783
36.
(5.3;60)
Y3030_VIBC3 (A5F637)
UPF0294 protein VC0395_A1830.
Chain: 1-281, pI: 5.28, Mw: 31895
VIUB_VIBC3 (A5F660)
Vibriobactin utilization protein viuB.
Chain: 1-271, pI: 5.33, Mw: 30515
VIUB_VIBCH (P0C6Q2)
Vibriobactin utilization protein viuB.
Chain: 1-271, pI: 5.33, Mw: 30515
Y4515_VIBVY (Q7MF05)
Putative phosphotransferase VVA0515.
Chain: 1-277, pI: 5.29, Mw: 31572
Y4006_VIBVU (Q8D7Y9)
Putative phosphotransferase VV2_0006.
Chain: 1-277, pI: 5.29, Mw: 31572
Y2238_VIBCH (Q9KPX4)
UPF0294 protein VC_2238.
Chain: 1-281, pI: 5.28, Mw: 31895
37.
(5.4;34)
PYRB_VIBS2 (P96174)
Aspartate carbamoyltransferase catalytic chain.
Chain: 2-310, pI: 5.44, Mw: 34288
Y3015_VIBPA (Q87KG2)
UPF0276 protein VP3015.
Chain: 1-288, pI: 5.36, Mw: 33257
RDGC_VIBPA (Q87S56)
Recombination-associated protein rdgC.
Chain: 1-304, pI: 5.36, Mw: 34355
HEM3_VIBCH (Q9KVM1)
Porphobilinogen deaminase.
Chain: 1-311, pI: 5.36, Mw: 33849
38.
(5.5,36)
TCPF_VIBC3 (A5F383)
Toxin coregulated pilus biosynthesis protein F.
Chain: 21-338, pI: 5.50, Mw: 35846
YGFZ_VIBC3 (A5F5F3)
tRNA-modifying protein ygfZ.
Chain: 1-323, pI: 5.49, Mw: 36109
TTCA_VIBC3 (A5F888)
tRNA 2-thiocytidine biosynthesis protein ttcA.
Chain: 1-310, pI: 5.51, Mw: 35123
TCPF_VIBCH (P0C6Q5)
Toxin coregulated pilus biosynthesis protein F.
Chain: 21-338, pI: 5.50, Mw: 35846
TTCA_VIBVY (Q7MKU7)
tRNA 2-thiocytidine biosynthesis protein ttcA.
Chain: 1-310, pI: 5.53, Mw: 35186
TTCA_VIBVU (Q8D9J0)
tRNA 2-thiocytidine biosynthesis protein ttcA.
Chain: 1-310, pI: 5.53, Mw: 35183
TTCA_VIBCH (Q9KS29)
tRNA 2-thiocytidine biosynthesis protein ttcA.
Chain: 1-310, pI: 5.53, Mw: 35121
39.
(5.7;36)
GCP_VIBC3 (A5F9E8)
Probable O-sialoglycoprotein endopeptidase.
Chain: 1-339, pI: 5.71, Mw: 36507
META_VIBPA (Q87NW7)
Homoserine O-succinyltransferase.
Chain: 1-313, pI: 5.69, Mw: 36272
PDXA_VIBVU (Q8DED3)
4-hydroxythreonine-4-phosphate dehydrogenase.
Chain: 1-328, pI: 5.70, Mw: 35492
40.
(5.8;40)
LUXP_VIBVY (Q7MD15)
Autoinducer 2-binding periplasmic protein luxP.
Chain: 14-366, pI: 5.82, Mw: 39961
FADA_VIBVY (Q7MQI0)
3-ketoacyl-CoA thiolase.
Chain: 1-387, pI: 5.78, Mw: 40848
FADA_VIBVU (Q8DDK5)
3-ketoacyl-CoA thiolase.
Chain: 1-387, pI: 5.78, Mw: 40866
41.
(6;43)
NAGA_VIBFU (P96166)
N-acetylglucosamine-6-phosphate deacetylase.
Chain: 1-399, pI: 6.00, Mw: 43122
LPXB_VIBPA (Q87MF0)
Lipid-A-disaccharide synthase.
Chain: 1-379, pI: 5.99, Mw: 42764
CSD_VIBCH (Q9KPQ7)
Probable cysteine desulfurase.
Chain: 1-404, pI: 6.07, Mw: 43439
CYSA_VIBCH (Q9KUI0)
Sulfate/thiosulfate import ATP-binding protein cysA.
Chain: 1-376, pI: 6.09, Mw: 42133
TRPB_VIBME (Q9RCE8)
Tryptophan synthase beta chain.
Chain: 1-391, pI: 5.96, Mw: 42295
42.
(5.8;58)
OPGG_VIBC3 (A5F1Q1)
Glucans biosynthesis protein G.
Chain: 36-545, pI: 5.83, Mw: 57070
OXAA_VIBVY (Q7MQK5)
Inner membrane protein oxaA.
Chain: 1-539, pI: 5.79, Mw: 60418
PYRG_VIBPA (Q87LP9)
CTP synthase.
Chain: 1-546, pI: 5.83, Mw: 60127
OXAA_VIBVU (Q8DDI2)
Inner membrane protein oxaA.
Chain: 1-539, pI: 5.79, Mw: 60432
Y1931_VIBCH (Q9KQR7)
UPF0061 protein VC_1931.
Chain: 1-489, pI: 5.83, Mw: 55630
OPGG_VIBCH (Q9KSG8)
Glucans biosynthesis protein G.
Chain: 36-545, pI: 5.83, Mw: 57070
43.
(5.8;81)
KATG_VIBC3 (A5F7X7)
Catalase-peroxidase.
Chain: 1-724, pI: 5.78, Mw: 80681
RELA_VIBSS (P55133)
GTP pyrophosphokinase.
Chain: 1-744, pI: 5.84, Mw: 84547
GLGB_VIBCH (Q9KNE8)
1,4-alpha-glucan-branching enzyme.
Chain: 1-666, pI: 5.76, Mw: 77096
KATG_VIBCH (Q9KRS6)
Catalase-peroxidase.
Chain: 1-724, pI: 5.78, Mw: 80651
44.
(5.6;70)
DXS_VIBVY (Q7MN49)
1-deoxy-D-xylulose-5-phosphate synthase.
Chain: 1-621, pI: 5.61, Mw: 67946
UBID_VIBVU (Q8DDP0)
3-octaprenyl-4-hydroxybenzoate carboxy- lyase.
Chain: 1-617, pI: 5.63, Mw: 69466
DXS_VIBVU (Q8DFA3)
1-deoxy-D-xylulose-5-phosphate synthase.
Chain: 1-621, pI: 5.61, Mw: 67959
45.
(5.7;93)
MUTS_VIBC3 (A5F9C4)
DNA mismatch repair protein mutS.
Chain: 1-862, pI: 5.71, Mw: 96327
MUTS_VIBVY (Q7MHR2)
DNA mismatch repair protein mutS.
Chain: 1-853, pI: 5.70, Mw: 94485
MUTS_VIBVU (Q8DC53)
DNA mismatch repair protein mutS.
Chain: 1-853, pI: 5.70, Mw: 94483
LUXQ_VIBCH (Q9KLK7)
Autoinducer 2 sensor kinase/phosphatase luxQ.
Chain: 1-857, pI: 5.74, Mw: 96930
MUTS_VIBCH (Q9KUI6)
DNA mismatch repair protein mutS.
Chain: 1-862, pI: 5.71, Mw: 96327
GYRB_VIBCH (Q9KVX3)
DNA gyrase subunit B.
Chain: 1-805, pI: 5.69, Mw: 89520
46.
(5.78;98)
LUXQ_VIBCH (Q9KLK7)
Autoinducer 2 sensor kinase/phosphatase luxQ.
Chain: 1-857, pI: 5.74, Mw: 96930
47.
(4.4;36)
OMPU_VIBC3 (A5F934)
Outer membrane protein U.
Chain: 22-341, pI: 4.48, Mw: 34657
ZIPA_VIBHB (A7MT68)
Cell division protein zipA homolog.
Chain: 1-323, pI: 4.33, Mw: 36041
OMPU_VIBCH (P0C6Q6)
Outer membrane protein U.
Chain: 22-341, pI: 4.41, Mw: 34656
ZIPA_VIBPA (Q87RJ5)
Cell division protein zipA homolog.
Chain: 1-316, pI: 4.45, Mw: 35292
OMPU_VIBVU (Q8DBX0)
Outer membrane protein U.
Chain: 22-340, pI: 4.32, Mw: 34756
ZIPA_VIBVU (Q8DFK4)
Cell division protein zipA homolog.
Chain: 1-311, pI: 4.50, Mw: 34665
11/18/2008
Protein Gel
Samples used:
| Sample |
Volume |
||
| 1 |
Anoxic 6 hours |
2.75 microL of the original sample |
|
| 2 |
Aerobic 6 hours |
2 microL of the original sample |
|
| 3 |
1/3 Anoxic 6 hours |
4.6 microL of the diluted sample |
|
| 4 |
1/3 Aerobic 6 hours |
4 microL of the diluted sampe |
|
| 5 |
1/10 Anoxic 6 hours |
1.4 microL of the diluted sample |
|
| 6 |
1/10 Aerobic 6 hours |
1.2 microL of the diluted sample |
Volumes of the samples were brought up to 10 microL with H2O. 10 microL of x2R reagent was added to each sample.
Samples were vortexed, boiled for 10 min, centrifuged for 2 min and run on a gel for 50 min, V=150
11/17/2008
Strips #4 & 6 (same sample, different protein amounts) from 11/10/2008 were reduced and alkylated according to Invitrogen protocol. Strips were loaded onto Novex tris-glycine SDS/PAGE gels, with positive end of the strip closest to the ladder. Gels were run @ 125V for 2.25 hrs.
Aerobic 1/3, 6hrs.
Aerobic 1/10, 6hrs.
11/13/2008
Strips #3 & 5 (same sample, different protein amounts) from 11/10/2008 were reduced and alkylated according to Invitrogen protocol. Strips were loaded onto Novex tris-glycine SDS/PAGE gels, with positive end of the strip closest to the ladder. Gels were run @ 125V for 2.25 hrs.
Staining of the 2 D gels. I used long(Basic) protocol.
The was one change to the protocol: gels were developed for 13 min instead of recommended 4-8 min.
1/3 Anoxic, 6 hours
1/10 anoxic, 6 hours
cDNA gel of the samples from the pH experiment #2.
Gel prepartion:
1. mix 1 g of aragose with 75 microL of TAE.
2. heat in the microwave for 3 min, ( mix after 1 min)
3. Add 7.5 microL of Ethilium bromide
4. Let a gel to solidify
Loading map:
| 1 |
100 bp ladder |
| 2 |
pH = 4 + Chitinase |
| 3 |
pH = 5 + Chitinase |
| 4 |
pH = 6 + Chitinase |
| 5 |
Unaltered media (pH = 6.72) + Chitinase |
| 6 |
pH = 7.13 + Chitinase |
| 7 |
pH = 8 + Chitinase |
| 8 |
pH = 9 + Chitinase |
| 9 |
H2O + Chitinase |
| 10 |
pH = 10 + Chitinase |
| 11 |
pH = 4 + Vtubi_16sV2 |
| 12 |
pH = 5 +Vtubi_16sV2 |
| 13 |
pH = 6 + Vtubi_16sV2 |
| 14 |
Unaltered media (pH = 6.72) +Vtubi_16sV2 |
| 15 |
pH = 7.13 + Vtubi_16sV2 |
| 16 |
pH = 8 + Vtubi_16sV2 |
| 17 |
pH = 9 + Vtubi_16sV2 |
| 18 |
pH =10 + Vtubi_16sV2 |
| 19 |
H2O + Vtubi_16sV2 |

11/11/2008 -
IEF was performed on strips from yesterday with the following parameters:
250V - 20 mins.
450V - 15 mins.
750V - 15 mins.
2000V - 60 mins.
Casette (with strips) was stored @ -80C.
Notes: Casette was repeatedly tapped to dislodge tiny airbubbles that had accumulated O/N before focusing. Run looked pretty good. Some very slight bubbling towards the "-" end of the strips.
NOTE: Strips discarded 12/17/2010 as part of lab clean up. -
11/10/2008
2D Gel -IPG Strips (pH 4-7) Rehydration
Volumes of the samples required for the gels
| Sample |
Volume ( microL) |
| Anoxic 6 hours |
2.75 microL |
| Aerobic 6 hours |
2 microL |
| 1/3 of Anoxic 6 hours |
2.75/ 3 = 0.916666microL |
| 1/3 of Aerobic hours |
2 / 3 = 0.66666666microL |
| 1/10 of Anoxic 6 hours |
2.75/ 10 = 0.275 microL |
| 1/10 of Aerobic 6 hours |
2/10 = 0.2 microL |
Concentration of the original samples:
Anoxic 6 hours---685 microg/mL
Aerobic 6 hours ----935 microg/mL
Dilution were made in the following way:
1. Anoxic 6 hours sample:
2 microL of anoxic sample was mixed with 8 microL of H2O. Using C1V1= C2V2 formular I calculated concentration of the diluted sample:
C of the original anoxic sample = 685 microg/mL
V original = 2 microL
V final = 10 microL
685 microg/ mL * 2 microL = C * 10 microL
C = 137 microg/mL
2. Aerobic 6 hours sample:
2 microL of aerobic sample was mixed with 10 microL of H2O. Using C1V1= C2V2 formular I calculated concentration of the diluted sample:
C of the original aerobic sample = 935 microg/mL
V of the original sample = 2 microL
V of the final sample = 12 microL
935 microg/ mL * 2 microL = C * 12 microL
C = 156 microg/ mL
Calculation of the amount of protein required for 1/3 and 1/10 samples:
1. 1/3 Anoxic 6 hours:
V= 0.916666 microL
C=685 microg/mL
Amount required= 0.916666 microL*685 microg/mL * 1 mL/1000 microL = 0.627916 microg
Volume of the diluted sample reuired = 0.627616 microg / 137 microg/ mL * 10^3 microL/ 1 mL = 4.6 microL
2. 1/10 Anoxic 6 hours:
V = 0.275 microL
Amount of the protein required = 0.275 microL * 685 microg/mL * 1 mL/ 1000 microL = 0.188 microg
Volume of the diluted sample required = 0.188 microg / 137 microg/ mL * 10^ 3 microL/ 1mL = 1.4 microL
3. 1/3 Aerobic 6 hours:
V =0.666666 microL
C = 935 microg/mL
Amount of the protein required = 935 microg/ mL * 0.666666 microL * 1 mL/ 1000 microg = 0.6233 microg
Volume of the diluted sample required = 0.6233 microg / 156 microg/ mL * 10^3 microL/ 1 mL = 4 micro L
4. 1/10 Aerobic 6 hours:
V = 0.2 microL
C = 935 microg/mL
Amoutn of the protein required = 935 microg/mL * 0.2 microL * 1 ml/1000 microL = 0.187 microg
Volume of the diluted sample required = 0.187 microg / 156 microg/mL * 10^3 microL/ 1 mL = 1. 2 microL
Tubes:
| Sample |
Volume |
|
| 1 |
Anoxic 6 hours |
2.75 microL of the original sample |
| 2 |
Aerobic 6 hours |
2 microL of the original sample |
| 3 |
1/3 Anoxic 6 hours |
4.6 microL of the diluted sample |
| 4 |
1/3 Aerobic 6 hours |
4 microL of the diluted sampe |
| 5 |
1/10 Anoxic 6 hours |
1.4 microL of the diluted sample |
| 6 |
1/10 Aerobic 6 hours |
1.2 microL of the diluted sample |
Master mix:
1. 2D solubilizer # 2 ------------------------141 *7 = 987 microL
2. 2 M DTT -----------------------------------------0.775*7 = 5.4 microL
3. Carrier Ampholytes---------------------------0.775 * 7 = 5.4 microL
4. Bromophenol Blue---------------------------0.1*7 =0.7 microL
142. 65 microL of the master mix was added to each microtube. H2O was added to get volume of each sample to 155 microL.
2M DTT solution was made by adding 0.0308 g of the DTT powder (MW=154.2 g) to 100 microL of H2O.
155microL of the sample was loaded into sample loading well, add strips with gel side up ( follow protocol on pages 22-23 of ZoomIPRunnerSystem manual)
Strips were incubated at the RT overnight.
Order of loading is the following:
| 1 |
Anoxic 6 hours |
|
| 2 |
Aerobic 6 hours |
|
| 3 |
1/3 Anoxic 6 hours |
|
| 4 |
1/3 Aerobic 6 hours |
|
| 5 |
1/10 Anoxic 6 hours |
|
| 6 |
1/10 Aerobic 6 hours |
11/6/2008
-
IEF was performed on the 4 strips from yesterday with the following parameters:
250V - 20 mins.
450V - 15 mins.
750V - 15 mins.
2000V - 60 mins.
Notes: Dye front looked good initially, but then seem to disperse as focusing went on. Possibly bubbling of the gels on the strips.
Strips were incubated with 10mL of sample reducing buffer for 15mins. w/shaking and then 10mL of sample alkylation buffer for 15mins. w/shaking.
Strips were removed (NOTE: Strip #1 still had the protective cover on it. Don't know how this will affect the sample.) and loaded into Invitrogen ZOOM 4-20% tris-glycine gels with the '+' end of the trips closest to the ladder. Strips were covered with 400uL 0.5% agarose (made with 1X tris-glycine running buffer) . Agarose was allowed to solidify for 10mins. 10uL of SeeBlue ladder was loaded into ladder lane. Gels were run 125V for ~2hrs.
Stained 2-D gels
Followed the protocol that Sam gave me
Gel #1
Gel #2
Gel #3
Gel #4
THINGS THAT I NEED TO DO;- erase this information after you do them
pH viability experiment - ( performed by Sam)
1. DNA gel
pH experiment that I performed
1. Do PCR with chitinase and Vtubi_16sV2 primers on the following samples
| VT grown in Marine Broth |
VT grown in Sea Water |
| pH 6.62, t= 5 min |
pH 7.57, t=5 min |
| pH 8.61, t=5 min |
pH 8.57, t= 5 min |
| pH 9.56, t= 5 min |
|
| pH 8.61, t= 30 min |
pH 7.57, t=30 min |
| pH 6.62, t= 30 min |
|
| pH 7.62, t=30 min |
|
| pH 7.62, t= 1 hour |
pH =7.57, t=1 hour |
| pH 8.81, t= 1 hour |
pH 9.56, t=1 hour |
| pH 6.62, t=1 hour |
|
| pH 6.62, t=2 hours |
pH 8.57, t= 2 hours |
| pH 8.61, t= 2 hours |
pH 9.56, t= 2 hours |
| pH 7.62, 2 hours |
pH 7. 57, t= 2 hours |
1. VT grown in Marine broth, pH 7.62, t= 5 min
2. VT grown in Sea H2O, pH 8.57, t=30 min
3. Vt grown in Sea H2O, pH 9.56, t= 30 min
4. VT grown in Sea H2O, pH 8.57, t= 1 hour
11/5/08
2D Gel-IPG Strip Rehydration
Volumes used:
1. Anoxic 6h---------------5.5 microL
2. Aerobic 6h-------------4 microL
3. Anoxic 6h-------------2.75 microL
4. Aerobic 6h-----------2 microL
Master mix
1. 2D #1 Solubilizer-----------------141*5=705 microL
2. 2M DTT -----------------------------0.775*5 = 3.875 microL
3. Carrier Ampholytes------------0.775* 5 = 3.875 microL
4. Bromophenol Blue-------------0.1*5 = 0.5 microL
5. Add H2O to get volume to 155 miroL
Making 2M DTT solution:
Volume = 100 microL
Momecular weight of DTT = 154.2 g
Mass of DTT = 154.2 g/mole * 1L/1000000microL * 100 microL * 2 mole/L = 0. 0308 g
Procedure:
1. 142. 65 microL of rehydrating buffer was added to each sample .
2. H2O was added to each sample to bring total volume of the sample to 155 microL
3.155microL of the sample was loaded into sample loading well, add strips with gel side up ( follow protocol on pages 22-23 of ZoomIPRunnerSystem manual)
Strips were incubated at the RT overnight.
11/04/2008
PCR of samples from V. tubiashii pH Viability Experiment ( description of experiment is in Sam's notebook)
Primers used:
1. Chitinase
2. Vtubi_16sV2
Samples:
| 1 |
pH = 4 + Chitinase |
|
| 2 |
pH = 5 + Chitinase |
|
| 3 |
pH = 6 + Chitinase |
|
| 4 |
Unaltered media (pH = 6.72) + Chitinase |
|
| 5 |
pH = 7.13 + Chitinase |
|
| 6 |
pH = 8 + Chitinase |
|
| 7 |
pH = 9 + Chitinase |
|
| 8 |
pH = 10 + Chitinase |
|
| 9 |
H2O + Chitinase |
|
| 10 |
pH = 4 + Vtubi_16sV2 |
|
| 11 |
pH = 5 +Vtubi_16sV2 |
|
| 12 |
pH = 6 + Vtubi_16sV2 |
|
| 13 |
Unaltered media (pH = 6.72) +Vtubi_16sV2 |
|
| 14 |
pH = 7.13 + Vtubi_16sV2 |
|
| 15 |
pH = 8 + Vtubi_16sV2 |
|
| 16 |
pH = 9 + Vtubi_16sV2 |
|
| 17 |
pH =10 + Vtubi_16sV2 |
|
| 18 |
H2O + Vtubi_16sV2 |
|
1. 2X Go-taq ------------12.5 *12 =150 micriL
2. Pf ------------------------0.5 *12 = 6 miroL
3. Pr------------------------0.5 * 12 = 6 microL
4. H2O--------------------10.5 *12 = 126 microL
Each tube contains 24 microL of appropriate master mix and 1 microL of cDNA.
10/31/08 Revers Transcription of the RNA samples isolated on 10/24/2008
Total amount of RNA in each sample is approximately 681.7 ng
| Sample |
Volume of the RNA used to get approximately 681.7 ng of RNA |
| marineB, pH 8.61, 30 min |
1.7 microL |
| marineB, pH 8.61, 5 min |
5 microL |
| marineB, pH 8.31, 2 hours |
2.2 microL |
| marineB, pH 7.62, 2 hours |
1.2 microl |
| mirineB, pH 7.62, 30 min |
1.6 |
| marineB pH 6.62, 30 min |
2.7 |
| sea H2O, pH 7.57, 5 min |
2 |
| sea H2O, pH 7.57, 30 min |
3 |
| sea H2O, pH 7.57, 1 hour |
1.9 |
| sea H2O, pH 8.57, 5 min |
1.6 |
| sea H2O, pH 9.56, 1 hour |
5 |
| sea H2O, pH 9.56, 5 min |
5 |
Master mix
1. 5X Buffer = 4*14 = 56 microL
2. dNTP = 8* 14 = 112 microL
3. AMV = 1* 14 = 14 microL
4. Oligo primers = 1*14 = 14 microL
5. H2O = 1*14 = 14 microL
15 miroL of the master mix was added to each tube, volume of each tube was adjusted to 20 microL with H2O.
10/27/08
PROTEIN GEL
Calculations:
1. anoxic sample:
C= 685 microg/mL
we need 15 microg, so 15 *1000/685 = 21. 9 microL of the sample should be used.
2. Aerobic sample:
C= 935 microg/mL
we need 15 microg, so 15*1000/935 = 16 microL of the sample should be used.
3. Desalten vt + oysters
C = 2.303 microg/ microL
we need 15 microg, so V = 15/2.303 = 6.51 microL
4.desalted vt +H2O sample
C = 1.186 microg/ microL
we need 15 microg, so V =15/ 1.186 = 12.65 microL
To each sample each volume of the x2R reagent was added. After this, I followed protocol provided on the website.
Loading map:
Each sample contains 15 mirograms of protein:
| Total Volume added into a well- proteins mixed with x2R, Ratio is 1:1 |
|
| 1. Ladder |
10 microL |
| 2. C |
20 microL |
| 4. VE |
20 microL |
| 6. Desalted vt + oysters |
13 miroL |
| 8. Desalted vt +H2O |
25.3 microL |
| 10. VT aerobic 6 h |
32 microL |
| 12. VT anox 6h |
43.8 miroL |

10/24/08
Isolation of the RNA from the pH experiment.
Spec analysis of the sample gave the following results:




10/21/2008 -
2D Gels - V.tubi anoxic 6hr vs. aerobic 6hr
IEF focusing was performed on all 6 strips from yesterday.
250V - 20 mins.
450V - 15 mins.
750V - 15 mins.
2000V - 60 mins.
NOTE: IEF stips looked gnarly; very uneven and not smooth like they normally appear after a good run.
Anoxic 6hr and aerobic 6hr samples were prepped and run on Invitrogen tris-glycine SDS/PAGE gels according to Invitrogen protocol. "Plus" side of strips are closest to the ladder. However, SDS/PAGE gels ran for a total time of 2hrs and 10mins for dye front to run to end of gel. All other samples were stored at -80C.
Vt Anoxia 6hrs.

Vt Aerobic 6hrs.

Results: Both gels seem to suffer from protein overload. This is supported by the large number of "halos" (reverse-stained spots) on both gels, particularly the Aerobic samples, which most certainly had more protein, despite the quantitation results. The horizontal streaking is also an indicator of protein levels being too high. Samples will be quantitated again to reassess protein amounts.
10/20/2008
Protein quantification by the Coomassie Assay:
Samples analyzed:
Desalted samples from 20081018 were analyzed with the Coomassie quantification assay.
Procedure:
1. 5microL of the diluent was mixed with 250microL of the Coomassie reagent
2. Incubated at RT for 10 min
3. Asborbace measured at 595 nm gave the following results:
| Sample |
Average A |
Concentration |
Volume used for Protein gel (15microg) |
| VT+ hem, 3 h |
0.173 |
2475.7 microg/mL |
6.1 microL |
| VT plate control, 3h |
0.184 |
2628.09 |
5.7 |
| VT anox, 3h |
0.197 |
2818.57 |
5.3 |
| VT anox, 6 h |
0.233 |
3323 |
4.5 |
| VT aerobic, 3h |
0.226 |
3232 |
4.6 |
| VT aerobic, 6h |
0.192 |
2742.38 |
5.5 |
Volume of all samples was adjusted to 15 microL by adding H2O
Rehydrating buffer
1. Zoom 2D protein solubilizer 128 microL
2. DTT 0.000239g
3. Ampholyte 0.8 microL
4. Bromophenol Blue 0.5 microL
5. H2O 10.7 microL
6.Lysate 15 microL
Total volume is 155 microL
DTT Calculation:
We need 0.7 microL of 2M DTT solution:
C1V1=C2V2
C1= 2M-from the protocol
V1=0.7 microL-- from the protocol
C2= concentration of DTT in 140 microL
V1=140 microL
2M*0.7 microL = C2 * 140 microL
C2 = 0.01 M
We know that formular weight of the DTT is 154.2 g ---from the lable, so taking into consideration that M is the same thing as mole/L, and that our final volume is 155 miroL, we have:
0.01 mole/L*154.2 g/mole*1L/1000000microL *155 microL = 0.000239 g of DTT is required.
Master mix:
1. Zoom 2D protein solubilizer =128 microL * 6.5 = 832 microL
2. DTT =0.000239g * 6.5 = 0.0016 g
3. Ampholyte = 0.8 microL * 6.5 = 5.2 microL
4. Bromophenol Blue = 0.5 microL * 6.5 = 3.25 microL
5. H2O =10.7 microL * 6.5 = 69. 55 microL
6.Lysate = 15 microL * 6.5 = 97.5 microL
Total volume is = 155 microL * 6.5 = 1007.5 microL
Procedure:
1. 144 microL of rehydrating buffer was added to each sample to bring total volume to 155microL.
2.155microL of the sample was loaded into sample loading well, add strips with gel side up ( follow protocol on pages 22-23 of ZoomIPRunnerSystem manual)
Strips were incubated at the RT overnight.
Stips were loaded in the following order:
1. VT + hem, 3h------------Put a black mark with the sharpie next to it
2. VT plate control
3. VT anox, 3hours
4. VT anox, 6 hours
5. VT aerobic, 3 hours
6. VT aerobic, 6 hours
10/17/2008
Aerobic/ Anaerobic Experiment
Samples were collected after 3 hours, and spected: Aarobic = 1.432, Aanaerobic=0.628
Each sample was split into two tubes, centrifuged for 15 min at 4,000 RPM
Put in -80C freezer.
Hemocytes Experiment
Samples were collected from the plates, centrifuged for 15 min at 4,000RPM
Liquid decanted, tubes placed in -80 freezer.
Plates were washed with 1ml of Tri reagent, liquid collected in 1ml tubes.
Summery of RNA concentration vs. pH for the V. tubiashii pH Viability Experiment

Maximum growth at pH 6.72. pH 9----Outlier??????? (maybe less bacteria was added to the sample, something went wrong
wrong when RNA extraction was performed,etc)
| pH |
Concentration |
| 4 |
174 |
| 5 |
183.11 |
| 6 |
1030 |
| 6.72 |
1612.78 |
| 7.13 |
1444.95 |
| 8 |
1142.78 |
| 9 |
521 |
| 10 |
1031.96 |
10/10/08
Spectr. analysis of the samples from 10/06/08
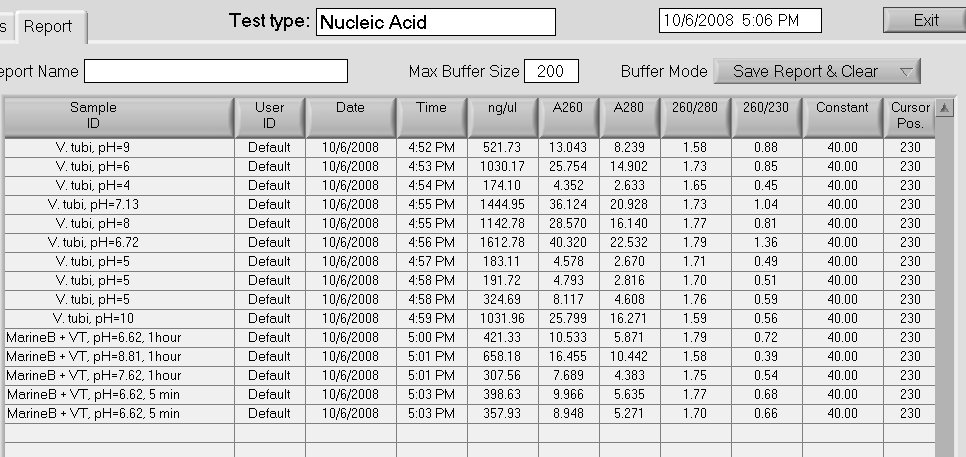
Reverse Transcription of the Samples
Amount of the RNA used for the reverse transcription:
SAmples 1-10-------915 ng
Samples 11-13-----1535 ng
10/06/08
Extraction of the RNA from the following samples:
1. V.tubi pH =6
2. V.tubi pH=7.13
3. V.tubi pH=6.72
4. V.tubi pH=9
5. V.tubi pH=8
6. V.tubi pH =10
7. V.tubi pH = 4
8. V tubi pH =5
9. MarineB+VT pH =6.62, 1hour
10. MarineB+VT pH =8.61, 1 hour
11. MarineB+VT pH=7.62, 1hour
12. MarineB + VT pH = 6.62, 1 hour
Samples 1-8 come from V. tubiashii pH Viability Experiment, samples 9-12 come from pH Experiment
9/26/08
repeated qPCR for samples from exposure and hemocyte challenge experiment with ftsz primes. Annealing temperature
was set to 50C and extension time to 1 minute.
Vt Marushchak
View SlideShare presentation or Upload your own.
9/24/08
cDNA Gel
PCR samples made by Sam on 9/23/08
Order of loading:
1. ladder 100bp, 15 microL
2. V.t + Auto Gigas, toxR, t=0
3. V.t + Auto Gigas, toxR, t=2
4. V.t + Auto Gigas, toxR, t=4
5. V.t + Auto Gigas, toxR, t=24
6. toxR +H2O
7. V.t + Auto Gigas, ompW, t=0
8. V.t + Auto Gigas, ompW, t=2
9. V.t + Auto Gigas, ompW, t=4
10. V.t + Auto Gigas, ompW, t=24
11. ompW + H2O
12. V.t + Auto Gigas, ftsz, t=0
13. V.t + Auto Gigas, ftsz, t= 2
14. V.t + Auto Gigas, ftsz, t=4
15. V.t + Auto Gigas, ftsz, t= 24
16. fitz +H2O
No bands are detected
9/22/08
Reverse Transcription of the following samples:
1. t=0, Vt + sterile sea water
2. t=0, Vt + live gigas
3. t=0, Vt + autoclaved gigas
4. t=0, gigas sea water
5. t=2, Vt + sterile sea water
6. t=2, Vt + live gigas
7. t=2, Vt + autoclaved gigas
8. t=2, gigas sea water
9. t=4, Vt + sterile sea water
10. t=4, Vt + live gigas
11. t=4, Vt + autoclaved gigas
12. t=4, gigas sea water
13. t=24, Vt+ sterile sea water
14. t=24, Vt + live gigas
15. t=24, Vt + autoclaved gigas
16. t=24, gigas sea H2O
17.Gigas plated hemos, C=67.9 ng/microL, Vused =5 microL
18. Gigas plated hemos +Vt (3h), C=0.125 microg/microL, V used=2.7 microL
19. Vt, 3 hours, 12C, C=0.104 microg/microL, Vused=3.3 microL
20. Gigas plated hemos + Vt supernatant, C=93.88 ng/microL, Vused=3.6 microL
Random promers were used
Calculations for Reverse Transcription: samples 1-16
I used formula C1V1=C2V2, to find volume of the RNA
Amount of the RNA used for reverse transcription is 5*64=320 ng
| Time |
Sample |
Concentration |
Volume |
| 0 |
Vt + sterile sea water |
82.25ng/microL |
3.9 |
| 0 |
Vt + live gigas |
0.160microg/microL |
2 |
| 0 |
Vt + autoclaved gigas |
0.179microg/microL |
1.8 |
| 0 |
gigas sea water |
0.133microg/microL |
2.4 |
| 2 hours |
Vt + sterile sea water |
0.248microg/microL |
1.3 |
| 2 h |
Vt + live gigas |
0.242 microg/microL |
1.3 |
| 2 h |
Vt + autoclaved gigas |
0.803 microg/microL |
dilution- see below |
| 2 h |
gigas sea water |
68.56 ng/microL |
4.7 |
| 4 h |
Vt + sterile sea wate |
0.254 microg/microL |
1.3 microL |
| 4 h |
Vt + live gigas |
0.261 microg/microL |
1.3 microL |
| 4 h |
Vt + autoclaved gigas |
1.456 microg/microL |
dilution-see below |
| 4 h |
gigas sea water |
64 ng/microL |
5 microL |
| 24 h |
Vt + sterile sea wate |
94.5 ng/ microL |
3.4 |
| 24 h |
Vt + live gigas |
0.208 microg/microL |
1.5 |
| 24 h |
Vt + autoclaved gigas |
0.208 microg/microL-sample was diluted after RNA extraction |
1.5 |
| 24 h |
gigas sea water |
87.98 ng/microL |
3.6 |
t =2 h , 1 microL of sample diluted with 11.5 microL of H2O, Cfinal = 64 ng/mL --------- 5 microL of diluted RNA used
t = 4 h, 1 microL of sample diluted with 21.7 microL of H2O, Cfinal = 64 ng/mL ----------5 microL of diluted RNA used
9/19/08
Expected size of the FliM product:
Forward primer: 343->362
Reverse primer: 871->849
Expected size = 871 - 343 = 528 bases
Minipreps- Plasmid DNA Purification
Samples used:
1. LAP
2. FST-L
3. COX
4. Decorin
Procedure:
1. Take 1 ml of bacteria cell suspension, centrifuge for 1 min, decant supernatant
2. Follow protocol on page 23 of QIAprep Miniprep Handbook
There were two changes to the protocol: step 7 was eliminated and 30 microL instead of 50 microL of the Buffer EB was used for elution.
Samples were analyzed and gave the following results:

9/16/08
Continuation of 2D Protein Gel
Isoelectric focusing (Protocol is on pages 24-29 of ZoomIPRunnerSystem manual)
Voltages:
| Voltage |
Dial on Power Supply |
Time |
| 200V |
5.5 |
20 minutes |
| 450V |
15 |
15 min |
| 750V |
26.5 |
15 min |
| 2000V |
68 |
30 min |
10 microL of the ladder was used
Gel #1 - Vt, no oyster
Gel #2 - Vt, autoclaved oysters
9/15/08
Protein quantification by the Coomassie Assay:
Samples analyzed:
1.VT + alive oysters, t=24
2. VT + autoclaved oysters, t =24
3. VT H2O, t=24
Procedure:
1. Pellets were thawed and resuspened in the 500 microL of Lysis buffer (pBAD)
2. Samples were frozen on dry ice and thawed at 42C. (Step was repeated 3 times)
3. Centrifuged at max speed for 1 min at -4C
4. Triton X-100, X-114 can interfere with the assay ( compatible C=0.125%). Concentration in our solution is 0.5%. To minimize the effect of the interfering substance, 50microL of diluent was diluted with 200microL of H2O. (Final concentration of the Triton X-100, X-114 =0.100%
5. 5microL of the diluent was mixed with 250microL of the Coomassie reagent
6. Incubated at RT for 10 min
7. Asborbace measured at 595 nm gave the following results:

Concentration of the proteins in diluted samples was calculated by averaging the absorbance and using equation:
C=((Aav-0.04)+0.0403)/0.00007
| VT + alive oysters, t=24 |
8,485.2 microg/mL |
| VT + autoclaved oysters, t=24 |
10,897.1 microg/mL |
| VT H2O, t=24 |
7,932.9 microg/mL |
| VT + alive oysters, t=24 |
42,426 microg/mL |
| VT + autoclaved oysters, t=24 |
54,485.5 microg/mL |
| VT H2O, t=24 |
32,664.5 microg/mL |
2D protein Gel
Samples used:1. VT + alive oysters, t=24
2. VT + autoclaved oysters, t=24
To get 15 microg of the protein in the sample,
1.8 microL of VT+alive oysters sample and 1.4 microL of VT + autoclaved oyster sample was used
Procedure
1. add appropriate volume of the rehydrating buffer to the sample to bring volume to 155microL
2. Load 155microL of the sample into sample loading well, add strips with gel side up ( follow protocol on pages 22-23 of ZoomIPRunnerSystem manual)
Strips were incubated at the RT overnight.
9/12/ 08 Chitinase and vthB look good on qPCR (however no big differences) FliM had no products.
suggestion: examine earlier timepoint. -  sr320 Sep 15, 2008
sr320 Sep 15, 2008
Friday Sam ran qPCR on subset with 3 primers, chitinase, vthB and FliM. -  sr320 Sep 14, 2008
sr320 Sep 14, 2008
Summery of the Exposure Experiment
This experiment was designed to examine the growth of the Vibrio tubiashii bacteria exposed to different treatments. Four tanks were set up at the room temperature. Three tanks received 4.435 X 10^11 Vibrio tubiashii bacteria (32ml bacteria/tank). Each tank was randomely allocated among three different treatment: tank 1 had sterile water, tank 2 had alive oysters and sea water, tank 3 had autoclaved oysters and sterile water. Fourth tank was set up as a control and contained alive oysters in sea water WITHOUT Vibrio tubiashii bacteria. Two 1 ml samples were taken from each tank for RNA/Protein analysis. Samples were drawn at the following time intervals: 0, 2 hours, 4 hours, 21 hours and 24 hours.
9/3/08
Flagella primers
Primers for FliM flagella protein were designed:
Vibrio FliM Rv GCGTGCTCAGGCATTTCAATC
Vibrio FliM Fw TGATTACCATGGAAGCGCGT
Protein Gel of some samples from pH and Oyster exposure experiments
Following samples were run on a protein gel on Friday:
1. Ladder
2. sea H2O, pH 7.57 after 30 min s
3. sea H2O, pH 8.57 after 30 min
4. sea H2O, pH 9.56 after 30 min
5. Sterile H2O + V. tubiashii, T=4 hours
6. Alive oysters + V. tubiashii, T= 4 hours
7. Autoclaved oysters + V. tubiashii, T=4 hours (darker)
8. Oysters H2O, T= 4 hour
9. Sterile H2O + V. tubiashii, T= 21 hours
10. Alive oysters + V. tubiashii, T= 21h
11. Autoclaved oysters + V.T., T = 21h (darker)
12. Oysters H2O, T = 21h
2 3 4 5 6 7 8 9 10 11 12
Analysis_ No dramatic difference in effect of pH after 30 min.
as expected sample with autoclaved oysters had more Vt thus more protein.
Based on the photos though it did not seem much higher than Vt grown in sterile seawater.-
8/29/08
Description of pH experiment
This experiment is designed to check responses of V. tubiashii to different pH values. 1 ml of V. tubiashii bacteria (C=1.386x10^10 bac/ml) was added to 19 mL of either marine broth or sea H2O of different pH.
Three different pH values were tested for marine broth and sea H2O. Initial pH of the marine broth and sea H2O was measure and gave the values of 7.62 and 8.57 respectively. 0.1 N HCl was used to decrease pH by approximately one unit and 0.1 NaOH was used to increase pH by 1 unit.
Thus, 1 ml of V. tubiashii was added in the tubes containing either marine broth or sea H2O with the following pH:
| Marine Broth |
Sea H2O |
| 7.62 |
8.57 |
| 6.62 |
7.57 |
| 8.61 |
9.56 |
All tubes were put on the shaker and temperature was set at 20C. 1 mL samples (2) were taken after 5 min, 30min, 1 hour, 2 hours and 24 hours.
-Roberts 8/28/08 11:29 AM
Final samples taken from oyster / Vt exp. includes 45 ml pellet-- soup also
All oysters- hemocytes and gill samples taken
-Roberts 8/28/08 8:02 AM
Sampled (2x 1ml) from 4tank oyster/Vt experiment
pellet- freeze
 |
| DSCF0057.JPG |
-Roberts 8/28/08 7:44 AM
10 ml left in tubes from ph experiment
spun down and froze
-Roberts 8/28/08 6:57 AM
Sampling (terminal) from pH experiment
6 treatments
taking 1 ml sample (2) from each
remaining liquid in 50 ml falcon also preserved.
 |
| Vt pH experiment by you. |
 |
| DSCF0054.JPG by you. |
-Roberts 8/27/08 1:20 PM
pulled second round of samples
-Roberts 8/27/08 11:28 AM
Treatment started.
4 tanks. Vt tanks received 4.435 X 10^11 bacteria (32ml bacteria/tank).
1 ml samples taken (2) from each tank for for RNA / Protein analysis
 |
| DSCF0044.JPG by you. |
 |
| DSCF0052.JPG by you. |
Steven 8/27/08
Today start a treatment.
Yesterday gel, VthB / chitinase might be only primers working consistently.
Interesting 16s absence.
Steven 8/26/08
Things on our minds: flagellin, changing pH
Tatyana 8/25/08
Set up PCR for V.tubiashii grown with clam foot and without foot.
Primers used:
1. Vtubi_16sV2
2. Vtubi_chitinase
3. Vtubi_VthB
PCR tubes:
| 1. V. tubiashii with foot-1 + Vtubi_16sV2 |
6. V. tubiashii with foot-1 + Vtubi_chitinase |
11. V. tubiashii with foot-1 + Vtubi_VthB |
| 2. V. tubiashii with foot-2 + Vtubi_16sV2 |
7. V.tubiashii with foot-2 + Vtubi_chitinase |
12. V. tubiashii with foot-2 + Vtubi_VthB |
| 3. V. tubiashii no foot-1 + Vtubi_16sV2 |
8. V.tubiashii no foot-1 + Vtubi_chitinase |
13. V. tubiashii no foot-1 + Vtubi_VthB |
| 4. V. tubiashii no foot-2 + Vtubi_16sV2 |
9. V.tubiashii no foot-2 + Vtubi_chitinase |
14. V. tubiashii no foot-2 + Vtubi_VthB |
| 5. H2O + Vtubi_16sV2 |
10. H2O + Vtubi_chitinase |
15. H2O + Vtubi_VthB |
Anealing tempereature is set to 55 C.
Tubes 31 and 32 from PCR of V. Tubiashii, V.Parahemoliticus, V. vulnificus cDNA experiment are placed in the PCR machine as well.
-Roberts 8/25/08 12:19 PM
23 pos (interpt) vthB - species specific
Tatyana 8/22/08
PCR of V. Tubiashii, V.Parahemoliticus, V. vulnificus cDNA.
I took 9 microL of cDNA and diluted it with 9microL of H2O to get enough cDNA. The following primers were used:
1. Vtubi_16sV2
2. Vrpos
3. Vphil_contig854
4. Vtubi_chitinase
5. L-tdh + R-tdh
6. Vtubi_VthB
7. Vspa24
8. VP 21,22
I set anealing temperature to 55 C.
Here is what is in the PCR tubes:
| 1. V. Parahemol. + Vtubi_16sV2 2. V. vulnificus +Vtubi_16sV2 3. V. tubiashii + Vtubi_16sV2 4. H2O + Vtubi_16sV2 5. V. Parahemol. + Vrpos 6. V. vulnificus + Vrpos 7. V. tubiashii + Vrpos 8. H2O + Vrpos 9. V. para + Vphil_contig854 10. V. vulnificus + Vphil_contig854 11. V. tubiashii + Vphil_contig854 12. H2O + Vphil_contig854 13. V. para + Vtubi_chitinase 14. V. vulnificus + Vtubi_chitinase 15. V. tubiashi + Vtubi_chitinase 16. H2O + Vtubi_chitinase |
17. V. para + L-tdh + R-tdh 18. V. vulfinicus + L-tdh + R-tdh 19. V. tubiashii + L-tdh + R-tdh 20. H2O + L-tdh + R-tdh 21. V. para + Vtubi_VthB 22. V. vulnificus + Vtubi_VthB 23. V. tubiashii + Vtubi_VthB 24. H2O + Vtubi_VthB 25. V. para + Vspa24 26. V. vulnificus + Vspa24 27. V. tubiashii + Vspa24 28. H2O + Vspa24 29. V. para + VP 21,22 30. V. vulnificus + VP 21,22 31. V. tubiashii + VP 21,22 32. H2O + VP21, 22 |
Because there are only 30 wells on the PCR block, I was not able to do PCR on the samples 31 and 32. I put them I my box.
Foot Experiment
Yesterday I isolated RNA from the V. tubiashii that was grown in the tube with the clam foot and in the tube without foot.
I did spectrophotometer analysis of prepared RNA and got the following concentrations:
| 1. V.Tubiashii +foot-1st tube |
1669.37 ng/ul |
| 2. V. Tubiashii +foot -2nd tube |
1399.70 ng/ ul |
| 3. V. tubiashii without foot - tube 1 |
351.01 ng/ ul |
| 4. V. tubiashii without foot - tube 2 |
251. 81 ng/ ul |
I diluted all the sample to get concentration to the 150 ng/ ul and then did spectr. analysis one more time. Here is the values that I got:
| 1. V.Tubiashii +foot-1st tube |
139 ng/ ul |
| 2. V. Tubiashii + foot - 2nd tube |
148 ng/ ul |
| 3. V. tubiashii without foot - tube 1 |
147 ng/ ul |
| 4. V. tubaishii without foot - tube 2 |
147 ng/ ulF |
Leftover of the V. tubiashii with foot and V. tubiashii without foot diluted in the TRI reagent are in the -80 freezer.
Today I did reverse transcription of these samples.
-Roberts 8/19/08 3:37 PM
1. QPX-SPX + HEMOS
2.SR4-SER-PRO + HEMOS
3. SER-PROB + HEMOS
4. Vtub-16s + hemos
5. R-TDH + hemos
6. VP21,22 + hemos
7. VRPos + hemos
8. QPX-SPX + sup. hemos + vt
9. QPX-SPX control
10. SR4-Ser-Pro control
11. SER-PROB control
12. Vtub-16s control
13. R-TDH control
14. VP21,22 control
15. VRPos control
16. only GoTaq + VRPos primers (I decided to test my GoTaq for contamination.)
-Roberts 8/19/08 10:34 AM
Summary experiments to date:
1) Simple temperature
2) Vt with clam foot
still need to get some RNA
3) Vt with hemocytes (liquid culture)
4) Vt with hemocytes (plated hemocytes)
SR4 SER PRO -- possibly only with hemocytes (plated and solution)
SerProB -- possibly only with hemocytes (plated and solution)
R-tdh -- signal only with non-adhered hemocytes
VPP22,21 (tdh) -- 1st four. just hemo, just Vt, with hemo
8/19/08 9:50 AM
Yesterday Tatyana ran some conventional PCR attempting to amplify serine protease product
RNA was extracted from Vp and Vv so generate cDNA for proper controls.
8/18/08 5:28 PM
8/18/08 11:41 AM
TTS system
see http://aem.asm.org/cgi/content/abstract/73/16/5183
ordered some spa24 primer that were designed by contiging Vp and Vc
-Roberts 8/18/08 11:29 AM
Serine Protease angle
see QPX presentation
SP in other Vibrio LINK
-Roberts 8/18/08 9:47 AM
summary- several SPs have signals.
Real-time from 8/8 experiment
Last night two poly-D lysine plates were seeded with 3.25mL Gigas hemos. Incubated 12C ~16hrs in the dark. 1mL Vt culture grown o/n at RT in MB(~28C, @ ~175RPM)
This AM, plates were gently washed with sterile sea water. 3 mL sea water added to plates after washing. 1 plate innocud with Vt (200uL), one with MB (200uL), and a third plate just had Vt added (200uL in 3mL sea water).
Incubated 3 hrs. at 12C in dark @ 50 RPM.
Plates with Vt = supe. collected, pelleted, remove supe, resuspend pellet in 1mL Tri-Reagent.
Plates with hemos = supe. removed, washed plates with 1mL Tri-Reagent.
RNA isolation by Tatyana.
SPB primers -- nothing
SR4 SER PRO -- possibly only with hemocytes (plated and solution) maybe increase anneal temp
SerProB -- possibly only with hemocytes (plated and solution)
Vtubi16s -- high signal with Vt. low signal in hemo withoug Vt. -increase temp
R-tdh -- signal only with non-adhered hemocytes
VPP22,21 (tdh) -- 1st four. just hemo, just Vt, with hemo
VRPOS (VP16s) -- nothing
Primers
SPB
F-CCAGGTGTTCTCTCGCTTTC
R-TCGTTGCACCAACTGTGATT
SR4 SER PRO
F-TGACCCACAAACAGACCTAGC
R-TGGGATAGCAAATCCAACTCC
Ser ProB
F-TTG ACG CAG TGG TAT GGA CGT AGT
R- ?????????????????????????????????
Vtubi_16sV2_F 466 GTGAGGTCGGTGGTTCAAGT
Vtubi_16sV2_R 465 ACCCCAAGACAACTGTGCTC
Second set of 16s primers maybe more specific??
VP22 464 GGGGATCCCTCAGTACAAAGCCTT
VP21 463 TGGTTGACATCCTACATGACTGTG
gene- tdh1 from parahemolyticus source (http://www.ncbi.nlm.nih.gov/pubmed/8126441 )
R-tdh 462 TGGAATAGAACCTTCATCTTCACC
L-tdh 461 GTAAAGGTGTCTGACTTTTTGAC
gene
VrposR 460 TCACCACGCAATGCTCTG
VrposF 459 GACAATGCGTCAGAGACG
Vtubi_16sF 456 CAGCCACACTGGAACTGAGA
Vtubi_16sR 455 GTTAGCCGGTGCTTCTTCTG
Vtubi_chitinase_rv 446 GTGCTGCCTTTCAGCTTACC
Vtubi_chitinase_fw 445 CGGACAACGGTATTCAGCTT
Vtubi_VthB_rv 444 TCACTGGGTCACCATTCTCA
Vtubi_VthB_fw 433 AAACCGATGCGCTCTACATC
-Roberts 8/18/08 6:40 AM
Draft Agenda for NOAA meeting on West coast shellfish-hatchery
larval production losses and solutions,
Olympia, WA
Wednesday, August 20, 10 am. Columbia Room,
Legislative Building, Washington State Capitol Campus
Conference call dial in: 1-888-387-8686 ID 2018106 #
Introductions
Background and status of current hatchery production
1. Vibrio tubiashi (Vt)i outbreaks at Netarts Bay 1998 and 2007: Hypothesis: Role of warmed seawater, upwelling, other factors in outbreaks Site differences that have been reported. RE
2. Hatchery production 2008: poor to suboptimal specific hatcheries, is Vt a primary factor now? Evidence from 2008. RE, BE, SC or AB
3. How big a current problem for hatchery production and the shellfish industry? (1) Taylor BE (2) WCSH SC and others wishing to contribute.
Current Research Activities
1. Prevalence and occurrence of Vt RE
2. WCSH seawater treatment system development AB, CL, SC
3. Bioassays of key factors, skimmer, UV, pH sod carb, other CL
4. Vibrio research - virulence factors and their genetic control CH
Research and Development Needs
1. Understanding and setting standards for seawater chemistry parameters for hatcheries
2. Developing water treatment, filtration, conditioning systems to meet these standards
3. Alkalinity and availability of essential minerals for larval shell formation in hatchery systems
4. Vt occurrence, pathology and control methods in hatcheries
5. Elucidate virulence factors of Vt and other pathogenic vibrios
6. Understand mechanism of expression of toxin and genes in Vt
7. Understand gene transfer in Vt
8. Vt in the environment, what triggers blooms and where is it
9. Other pathogenic vibrios - mechanisms of virulence (same questions as for Vt)
10. Understanding and standardized ways to measure bloom degradation products, indices and toxicity in estuarine and other hatchery environments
What are the Research Needs now?
Opportunities for funding for control measures and research…
-Roberts 8/18/08 6:26 AM
a few weeks ago an OMP primer pair produced a band (need more details)
-Roberts 8/15/08 12:12 PM
The underlying premise is that we need to know more about the basic physiology of Vt in order to develop measures to protect shellfish resources.
One means by which to do this is to look at differences in gene expression under different conditions. Conditions, being for example, alterations in: temperature, salinity, oxygen, host tissue. What genes are important to look at? Conventional comparative gene fishing techniques are not good options as most depend on polyA regions. Several genes have been identified including several that are implicated in virulence. To date there are 31 nucleotide entries in NCBI for Vt. These are obvious choices, however if samples are from sources where other Vibrios are present, effort has to be made to insure the lack of cross-reactivity with other organisms.
A second avenue would be to take a comparative approach using the complete genome sequence from V parahemolyticus and V vulnificus. Several tools are available for this including whole genome comparison where homologus genes are identified. This and other similar data can be found on the genome project page (example). For this approach you are basically flying blind not sure if 1) the gene is relavant or 2) if there is sufficient homology.
The techniques discussed so far are "old-fashion" PCR based. Another option would be to once again exploit the other Vibrios, but use a micro array platform, again relying on the assumption there is sufficient homology. Relative comparisons should be simple however.
Protein expression is another viable route. More on that later..
-Roberts 8/15/08 7:54 AM
16s PCR was done on earlier exp (see graphs)
concerned of overiding factors such as cross reactivity of primers.
-Roberts 8/8/08 10:26 AM
Last night two poly-D lysine plates were seeded with 3.25mL Gigas hemos. Incubated 12C ~16hrs in the dark. 1mL Vt culture grown o/n at RT in MB(~28C, @ ~175RPM)
This AM, plates were gently washed with sterile sea water. 3 mL sea water added to plates after washing. 1 plate innocud with Vt (200uL), one with MB (200uL), and a third plate just had Vt added (200uL in 3mL sea water).
Incubated 3 hrs. at 12C in dark @ 50 RPM.
Plates with Vt = supe. collected, pelleted, remove supe, resuspend pellet in 1mL Tri-Reagent.
Plates with hemos = supe. removed, washed plates with 1mL Tri-Reagent.
RNA isolation by Tatyana.
-Roberts 8/6/08 1:06 PM
note: pull-"Targets include genes associated with general metabolism (dsdA, Makino et al. 2003), adaptation to environmental stress (rpoS (alternate sigma factor of RNA polymerase) Vasudevan and Venkitanarayanan 2006, Coutard et al. 2007), lateral flagella synthesis (lafK, Merino et al. 2006), quorum sensing and virulence (opaR (luxR homolog) Henke and Bassler 2004), virulence (tdh2, Coutard et al. 2007; spa24 (component of TTSS system), Coutard et al. 2007). The housekeeping gene pvsA will be used for normalizing expression levels as described in Coutard et al 2007. "
-Roberts 8/6/08 9:57 AM
below repeated, still smear. though different size bands evident
possibly protease
4 day Vt growth with and without Mm foot tissue. with foot smear (protease?)
*note Vt data not normalized
Performed seed experiment at 18
-Roberts 7/22/08 10:28 AM
Culture Vt at 12 and 21
Reverse transcribed.
Reverse transcribed next day by Sam with equal starting RNA.
SAFS Seed Proposal:
Development of tools to monitor and predict outbreaks of Vibrio tubiashii
Carolyn Friedman and Steven Roberts
Current state of knowledge
Background Oyster production is important to the US both economically and socially. Pacific Crassostrea gigas and eastern C. virginica oysters are the two key culture species within the US with production of Pacific oysters estimated at 89,323,000 lbs (40,601 tonnes) in 2004 and eastern oysters at ~4,000 tonnes (WRAC, 2004; FAO 2006). Washington state produced a majority (~87% or 77,000,000 lbs) of US Pacific oysters and California produces most of the remaining Pacific oysters (7,000,000 lbs). In addition to harvest-sized product, 35 million Pacific oyster seed were produced in 1995 (WRAC 2004) and production has increased since that time.
Oyster Mortality Losses of oysters associated with disease has increased over the past few decades with expansion of known disease agents (such as MSX and Dermo; Fig. 1B) and the introduction of emerging diseases such as that caused by the oyster herpes virus (OsHV) and Vibrio tubiashii. The impact of diseases on wild and cultured oysters is considered a leading impediment to the oyster industry within the US and subsequently affecting the health of some of key estuaries such as the Chesapeake Bay (Andrews 1988, Burreson et al. 1996, Ragone Calvo et al. 2003). Economic impacts of diseases have hampered the sustainability of oyster culture in several US areas including the mid-Atlantic, Gulf and west coasts. Locally, the hatcheries along the west cost of the US have been impacted by vibriosis (disease caused by members of the genus Vibrio, including V. tubiashii) resulting in a lack of seed for many farms and partial closure of at least one hatchery (Jonathan Davis, pers. comm.). In fact, vibriosis caused by a variety of members of the genus Vibrio are pathogenic to a wide variety of marine invertebrates. Although larvae are most commonly affected, adult shellfish may also be susceptible (e.g. mature abalone in France (Nichols et al. 2002) and manila clams in Europe (see Paillard 2004)
Vibriosis (also called bacillary necrosis) was first described by Tubiash et al. in 1965 and was characterized by swarming of bacteria in and around oyster larvae, which stopped swimming, had empty guts and experienced high losses. This disease bacillary necrosis was further described by Brown (1973) who suspected the involvement of a toxin due to the rapid mortality of oyster larvae within 18 h of exposure. Two major toxins have been described in association with bacillary necrosis, one degrades connective tissues and the second is heat stable and ciliostatic, resulting in cessation of swimming (Nottage et al. 1989). Different host species are know to have varying degrees of susceptibility to specific strains of Vibrio; when challenged with V. alginolyticus scallops and pen shells were more susceptible than were Pacific oysters (Luna-Gonzalez et al. 2002). Recently, V. tubiashii has reemerged as a deadly oyster pathogen along the Pacific coast of the US. Current diagnosis is dependent upon culture methods (Elston et al., pers. comm.). Creating molecular diagnostic methods to complement current culture methods will enhance the capability of diagnosing this pathogen and will also enable the application of these tools to research questions. Recently, Hasegawa (2008) demonstrated that an extracellular metalloprotease is an important virulence factor this this bacterium. Such as gene would be an ideal target for a molecular test. Furthermore, developing techniques to assess how changes in the the environment influence V. tubiashii physiology will assist in predicting outbreaks and improving hatchery operating practices.
The specific objectives of the current proposal are to 1) develop an improved diagnostic assay to detect V. tubiashii and 2) develop gene expression assays to be to assess how environmental factors influence pathogen physiology.
Methods
We propose to develop conventional and real-time PCR tests for V. tubiashii. The 16S rDNA gene and metalloprotease genes of all V. tubiashi strains and representative related bacteria in GenBank will be aligned using ClustalW. We will select primers (using Primer 3 software) unique to our target bacterium and may also select primers general to this genus for later use to better understand bloom dynamics and relative concentrations for disease development. Primers will be tested using established methods (e.g. Vadopalas et al. 2005, Lyons et al. 2006). We will establish analytical and, as possible, diagnostic sensitivity and specificity according to methods outlined by OIE (2007). We will compare the molecular test to established culture methods currently employed using pure bacterial cultures as well as samples from local hatcheries and seed sources (e.g. Taylor Resources and Hog Island Oyster Company).
In order to determine the abiotic and biotic effects on V. tubiashi physiology related to growth rates, change in behavior (eg swarmiing), and virulence, real-time quantitative RT-PCR assays will be developed that will target expressed genes. Potential targets for the assay include genes associated with general metabolism (dsdA, Makino et al. 2003), adaptation to environmental stress (rpoS (alternate sigma factor of RNA polymerase) Vasudevan and Venkitanarayanan 2006, Coutard et al. 2007), lateral flagella synthesis (lafK, Merino et al. 2006), quorum sensing and virulence (opaR (luxR homolog) Henke and Bassler 2004), virulence (vtpA / vtpB, Hasegawa et al 2008; toxR Beaubrun et al 2008; tdh2, Coutard et al. 2007; spa24 (component of TTSS system), Coutard et al. 2007). RT-PCR reactions will be carried and analyzed as described above except RNA will be the template. Once assays have been developed and validated laboratory based experiments will be carried out to determine how factors such as temperature and salinity influence expression of certain genes. This will in turn give us information on how the physiology of the pathogen is altered under changing conditions.
Application for future
This research will provide important tools and preliminary data to garner future funds from Washington Sea Grant and/or the National Science Foundation (e.g. Ecology of Infectious Disease rfp). Without preliminary data and ready tools, obtaining funds and helping understand the disease dynamics will not be possible. In addition, the success of our west coast bivalve mollusc industry depends on being able to manage this bacterium. Having a reliable, rapid test as well as information on how environmental factors influence physiology will benefit the industry and researchers. In addition, V. tubiashi has also emerged in France (T. Renault, IFREMER, and S. Huchette, France Haliotis, pers. comm.), which will provide international collaborative opportunities.
Budget
References
Beaubrun JJ, Kothary MH, Curtis SK, Flores NC, Eribo BE, Tall BD. 2008 Isolation and characterization of Vibrio tubiashii outer membrane proteins and determination of a toxR homolog. Applied and Environmental Microbiology vol. 74 (3) pp. 907-11
Coutard F., Lozach S., Pommepuy M., Hervio-Heath D. 2007. Real-time reverse transcription-PCR for transcriptional expression analysis of virulence and housekeeping genes in viable but nonculturable Vibrio parahaemolyticus after recovery of culturability. Applied and Environmental Microbiology. 73(16): 5183-5189.
FAO. 2006
Hasegawa H, Lind EJ, Boin MA, Häse CC. 2008. The extracellular metalloprotease of Vibrio tubiashii is a major virulence factor for pacific oyster (Crassostrea gigas) larvae. Applied and Environmental Microbiology (2008) vol. 74 (13) pp. 4101-10
Henke J.M. & Bassler B.L. 2004. Quorum sensing regulates Type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. Journal of Bacteriology. 186(12): 3794-3805.
Lyons MM, Smolowitz R, Dungan C, Roberts SB. 2006. Development of a real-time quantitative PCR assay for the hard clam pathogen, Quahog Parasite Unknown (QPX). Diseases of Aquatic Organisms. 72(1):45-52
Makino K., Oshima K., Kurokawa K., Yokoyama K., Uda T., Tagomori K., Iijima Y., Najima M., Nakano M., Yamashita A., Kubota Y., Kimura S., Yasunaga T., Honda T., Shinagawa H., Hattori M., Iida T., 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet. 361: 743-749
Merino S., Shawe J., Tomas J.M. 2006. Bacterial lateral flagella: an inducible flagella system. FEMS Microbiological Letters 263: 127-135.
Nicolas JL, Basuyaux O, Mazurie J, Thebault A. 2002. Vibrio carchariae, a pathogen of the abalone Haliotis tuberculata. Diseases of Aquatic Organisms 50:35-43.
Paillard, C. 2004. a short-review of brown ring disease, a vibriosis affecting Ruditapes philippinarum and Ruditapes decussatus. Aquat. living Recourc. 17:467-475.
Vasudevan P., & Venkitanarayanan K. 2006. Role of the rpoS gene in the survival of Vibrio parahaemolyticus in artificial seawater and fish homogentate. Journal of Food Protection. 69(6): 1438-1442.
WRAC. 2004 Aquaculture production from 1994 to present.// http://www.fish.washington.edu/wrac/images/part2.pdf
LINKS**
http://seagrant.oregonstate.edu/blogs/h2onc/2008/06/09/vibrio-tubiashii-hypoxia-and-the-oysters-on-your-plat